Advancement in non-destructive methodologies for the determination of meat fraud
DOI:
https://doi.org/10.18686/fnc333Keywords:
food fraud; nondestructive methods; NIR spectroscopy; machine learning algorithm; linear regressionAbstract
Food fraud is a new term worldwide and can involve many different stages of the production process. It affects safety, quality, consumer acceptance, and profitability. Food fraud assessment methods need to be very precise and reliable. Most animal-origin foods, including milk, dairy products, meat and meat products, eggs, fish, and fisheries goods, are vulnerable to food fraud. Identifying any adulteration in them is essential to stop unfair competition and protect consumer rights. Due to financial benefits, meat and meat products are vulnerable to various forms of adulteration. The meat business is transitioning from laborious and time-consuming analytical procedures to quick, non-invasive, non-destructive, repeatable, and trustworthy analytical technologies. This reviews precision analytical methods like near-infrared (NIR) spectroscopy and machine learning algorithms, linear regression, principal component analysis, etc., for detecting food fraud in meat and meat products.
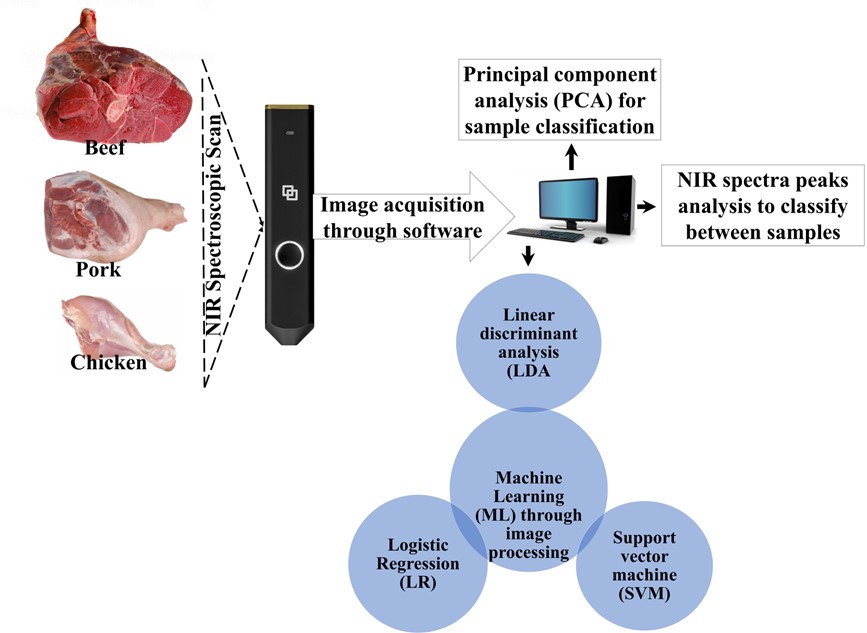
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Author(s)

This work is licensed under a Creative Commons Attribution 4.0 International License.
References
1. Karami H, Kamruzzaman M, Covington JA, et al. Advanced evaluation techniques: Gas sensor networks, machine learning, and chemometrics for fraud detection in plant and animal products. Sensors and Actuators A: Physical. 2024; 370: 115192. doi: 10.1016/j.sna.2024.115192 DOI: https://doi.org/10.1016/j.sna.2024.115192
2. Seesaard T, Goel N, Kumar M, et al. Advances in gas sensors and electronic nose technologies for agricultural cycle applications. Computers and Electronics in Agriculture. 2022; 193: 106673. doi: 10.1016/j.compag.2021.106673 DOI: https://doi.org/10.1016/j.compag.2021.106673
3. Abbas O, Zadravec M, Baeten V, et al. Analytical methods used for the authentication of food of animal origin. Food Chemistry. 2018; 246: 6-17. doi: 10.1016/j.foodchem.2017.11.007 DOI: https://doi.org/10.1016/j.foodchem.2017.11.007
4. Ballin NZ, Lametsch R. Analytical methods for authentication of fresh vs. thawed meat – A review. Meat Science. 2008; 80(2): 151-158. doi: 10.1016/j.meatsci.2007.12.024 DOI: https://doi.org/10.1016/j.meatsci.2007.12.024
5. Alamprese C, Amigo JM, Casiraghi E, et al. Identification and quantification of turkey meat adulteration in fresh, frozen-thawed and cooked minced beef by FT-NIR spectroscopy and chemometrics. Meat Science. 2016; 121: 175-181. doi: 10.1016/j.meatsci.2016.06.018 DOI: https://doi.org/10.1016/j.meatsci.2016.06.018
6. Ivanova I, Ivanov G, Shikov V, et al. Analytical Method for Differentiation of Chilled and Frozen-Thawed Chicken Meat. Acta Universitatis Cibiniensis Series E: Food Technology. 2014; 18(2): 43-53. doi: 10.2478/aucft-2014-0013 DOI: https://doi.org/10.2478/aucft-2014-0013
7. Alam AN, Hashem M, Matar A, et al. Cutting edge technologies for the evaluation of Plant-based food and meat Quality: A Comprehensive review. Meat Research. 2024; 4(1). doi: 10.55002/mr.4.1.79 DOI: https://doi.org/10.55002/mr.4.1.79
8. Barbin D, Elmasry G, Sun DW, et al. Near-infrared hyperspectral imaging for grading and classification of pork. Meat Science. 2012; 90(1): 259-268. doi: 10.1016/j.meatsci.2011.07.011 DOI: https://doi.org/10.1016/j.meatsci.2011.07.011
9. Cozzolino D, Murray I. Identification of animal meat muscles by visible and near infrared reflectance spectroscopy. LWT - Food Science and Technology. 2004; 37(4): 447-452. doi: 10.1016/j.lwt.2003.10.013 DOI: https://doi.org/10.1016/j.lwt.2003.10.013
10. Prieto N, Andrés S, Giráldez FJ, et al. Discrimination of adult steers (oxen) and young cattle ground meat samples by near infrared reflectance spectroscopy (NIRS). Meat Science. 2008; 79(1): 198-201. doi: 10.1016/j.meatsci.2007.08.001 DOI: https://doi.org/10.1016/j.meatsci.2007.08.001
11. Hashem M, Islam M, Hossain M, et al. Prediction of chevon quality through near infrared spectroscopy and multivariate analyses. Meat Research. 2022a; 2(6). doi: 10.55002/mr.2.6.37 DOI: https://doi.org/10.55002/mr.2.6.37
12. Wu D, Chen X, Shi P, et al. Determination of α-linolenic acid and linoleic acid in edible oils using near-infrared spectroscopy improved by wavelet transform and uninformative variable elimination. Analytica Chimica Acta. 2009; 634(2): 166-171. doi: 10.1016/j.aca.2008.12.024 DOI: https://doi.org/10.1016/j.aca.2008.12.024
13. Hashem M, Morshed M, Khan M, et al. Prediction of chicken meatball quality through NIR spectroscopy and multivariate analysis. Meat Research. 2022b; 2(5). doi: 10.55002/mr.2.5.34 DOI: https://doi.org/10.55002/mr.2.5.34
14. Prieto N, Pawluczyk O, Dugan MER, et al. A Review of the Principles and Applications of Near-Infrared Spectroscopy to Characterize Meat, Fat, and Meat Products. Applied Spectroscopy. 2017; 71(7): 1403-1426. doi: 10.1177/0003702817709299 DOI: https://doi.org/10.1177/0003702817709299
15. Thyholt K, Isaksson T. Differentiation of frozen and unfrozen beef using near-infrared spectroscopy. Journal of the Science of Food and Agriculture. 1997; 73(4): 525-532. doi: 10.1002/(SICI)1097-0010(199704)73:4<525::AID-JSFA767>3.0.CO;2-C DOI: https://doi.org/10.1002/(SICI)1097-0010(199704)73:4<525::AID-JSFA767>3.0.CO;2-C
16. Parastar H, van Kollenburg G, Weesepoel Y, et al. Integration of handheld NIR and machine learning to “Measure & Monitor” chicken meat authenticity. Food Control. 2020; 112: 107149. doi: 10.1016/j.foodcont.2020.107149 DOI: https://doi.org/10.1016/j.foodcont.2020.107149
17. Eui JM, Youngsik K, Yu X, et al. Evaluation of salmon, tuna, and beef freshness using a portable spectrometer. Sensors. 2020; 20(15): 4299. DOI: https://doi.org/10.3390/s20154299
18. Nolasco PIM, Badaró AT, Barbon S, et al. Classification of Chicken Parts Using a Portable Near-Infrared (NIR) Spectrophotometer and Machine Learning. Applied Spectroscopy. 2018; 72(12): 1774-1780. doi: 10.1177/0003702818788878 DOI: https://doi.org/10.1177/0003702818788878
19. Kamruzzaman M, ElMasry G, Sun DW, et al. Application of NIR hyperspectral imaging for discrimination of lamb muscles. Journal of Food Engineering. 2011; 104(3): 332-340. doi: 10.1016/j.jfoodeng.2010.12.024 DOI: https://doi.org/10.1016/j.jfoodeng.2010.12.024
20. Sanz JA, Fernandes AM, Barrenechea E, et al. Lamb muscle discrimination using hyperspectral imaging: Comparison of various machine learning algorithms. Journal of Food Engineering. 2016; 174: 92-100. doi: 10.1016/j.jfoodeng.2015.11.024 DOI: https://doi.org/10.1016/j.jfoodeng.2015.11.024
21. Alomar D, Gallo C, Castaneda M, Fuchslocher R. Chemical and discriminant analysis of bovine meat by near infrared reflectance spectroscopy (NIRS). Meat Science. 2003; 63(4): 441-450. doi: 10.1016/S0309-1740(02)00101-8 DOI: https://doi.org/10.1016/S0309-1740(02)00101-8
22. Al-Sarayreh M, M. Reis M, Qi Yan W, Klette R. Detection of red-meat adulteration by deep spectral–spatial features in hyperspectral images. Journal of Imaging. 2018 May 3; 4(5): 63. https://doi.org/10.3390/jimaging4050063 DOI: https://doi.org/10.3390/jimaging4050063
23. Sanchez PDC, Arogancia HBT, Boyles KM, et al. Emerging nondestructive techniques for the quality and safety evaluation of pork and beef: Recent advances, challenges, and future perspectives. Applied Food Research. 2022; 2(2): 100147. doi: 10.1016/j.afres.2022.100147 DOI: https://doi.org/10.1016/j.afres.2022.100147
24. Kamruzzaman M, ElMasry G, Sun DW, et al. Non-destructive assessment of instrumental and sensory tenderness of lamb meat using NIR hyperspectral imaging. Food Chemistry. 2013; 141(1): 389-396. doi: 10.1016/j.foodchem.2013.02.094 DOI: https://doi.org/10.1016/j.foodchem.2013.02.094
25. Slosarz P, Stanisz M, Boniecki P, et al. Artificial neural network analysis of ultrasound image for the estimation of intramuscular fat content in lamb muscle. African Journal of Biotechnology. 2011; 10(55): 11792.
26. Alaiz-Rodriguez R, Parnell AC. A Machine Learning Approach for Lamb Meat Quality Assessment Using FTIR Spectra. IEEE Access. 2020; 8: 52385-52394. doi: 10.1109/access.2020.2974623 DOI: https://doi.org/10.1109/ACCESS.2020.2974623
27. Fisher RA. The use of multiple measurements in taxonomic problems. Annals of Eugenics. 1936; 7(2): 179-188. doi: 10.1111/j.1469-1809.1936.tb02137.x DOI: https://doi.org/10.1111/j.1469-1809.1936.tb02137.x
28. Hao J, Ho TK. Machine Learning Made Easy: A Review of Scikit-learn Package in Python Programming Language. Journal of Educational and Behavioral Statistics. 2019; 44(3): 348-361. doi: 10.3102/1076998619832248 DOI: https://doi.org/10.3102/1076998619832248
29. Raschka S, Patterson J, Nolet C. Machine Learning in Python: Main Developments and Technology Trends in Data Science, Machine Learning, and Artificial Intelligence. Information. 2020; 11(4): 193. doi: 10.3390/info11040193 DOI: https://doi.org/10.3390/info11040193
30. Taspinar YS, Koklu M, Altin M. Classification of flame extinction based on acoustic oscillations using artificial intelligence methods. Case Studies in Thermal Engineering. 2021; 28: 101561. doi: 10.1016/j.csite.2021.101561 DOI: https://doi.org/10.1016/j.csite.2021.101561
31. Sabanci K, Aslan MF, Slavova V, et al. The Use of Fluorescence Spectroscopic Data and Machine-Learning Algorithms to Discriminate Red Onion Cultivar and Breeding Line. Agriculture. 2022; 12(10): 1652. doi: 10.3390/agriculture12101652 DOI: https://doi.org/10.3390/agriculture12101652
32. Ropelewska E, Slavova V, Sabanci K, et al. Discrimination of onion subjected to drought and normal watering mode based on fluorescence spectroscopic data. Computers and Electronics in Agriculture. 2022; 196: 106916. doi: 10.1016/j.compag.2022.106916 DOI: https://doi.org/10.1016/j.compag.2022.106916
33. Badillo S, Banfai B, Birzele F, et al. An Introduction to Machine Learning. Clinical Pharmacology & Therapeutics. 2020; 107(4): 871-885. doi: 10.1002/cpt.1796 DOI: https://doi.org/10.1002/cpt.1796
34. Tharwat A, Gaber T, Ibrahim A, et al. Linear discriminant analysis: A detailed tutorial. AI Communications. 2017; 30(2): 169-190. doi: 10.3233/aic-170729 DOI: https://doi.org/10.3233/AIC-170729
35. Dumalisile P, Manley M, Hoffman L, et al. Discriminating muscle type of selected game species using near infrared (NIR) spectroscopy. Food Control. 2020; 110: 106981. doi: 10.1016/j.foodcont.2019.106981 DOI: https://doi.org/10.1016/j.foodcont.2019.106981
36. Cessie SL, Houwelingen JCV. Ridge Estimators in Logistic Regression. Applied Statistics. 1992; 41(1): 191. doi: 10.2307/2347628 DOI: https://doi.org/10.2307/2347628
37. Breiman L. Random forests. Machine Learning. 2001; 45(1): 5-32. doi: 10.1023/A:1010933404324 DOI: https://doi.org/10.1023/A:1010933404324
38. Kircali AS, Shi JK, Yao X, et al. Predicting the Textural Properties of Plant-Based Meat Analogs with Machine Learning. Foods. 2023; 12(2): 344. doi: 10.3390/foods12020344 DOI: https://doi.org/10.3390/foods12020344
39. Capron X, Smeyers-Verbeke J, Massart DL. Multivariate determination of the geographical origin of wines from four different countries. Food Chemistry. 2007; 101(4): 1585–1597. DOI: https://doi.org/10.1016/j.foodchem.2006.04.019
40. Rousseau R, Govaerts B, Verleysen M, et al. Comparison of some chemometric tools for metabonomics biomarker identification. Chemometrics and Intelligent Laboratory Systems. 2008; 91(1): 54-66. doi: 10.1016/j.chemolab.2007.06.008 DOI: https://doi.org/10.1016/j.chemolab.2007.06.008
41. Mutihac L, Mutihac R. Mining in chemometrics. Analytica Chimica Acta. 2008; 612(1): 1-18. doi: 10.1016/j.aca.2008.02.025 DOI: https://doi.org/10.1016/j.aca.2008.02.025
42. Tian F, Yang L, Lv F, et al. Predicting liquid chromatographic retention times of peptides from the Drosophila melanogaster proteome by machine learning approaches. Analytica Chimica Acta. 2009; 644(1-2): 10-16. doi: 10.1016/j.aca.2009.04.010 DOI: https://doi.org/10.1016/j.aca.2009.04.010
43. Hastie T, Tibshiranii R, Friedman J. The elements of statistical learning: data mining, inference, and prediction, 2nd ed. Springer; 2009.
44. Frank IE, Lanteri S. Classification models: discriminant analysis, SIMCA, CART. Chemometrics and Intelligent Laboratory Systems. 1989; 5(3): 247-256. doi: 10.1016/0169-7439(89)80052-8 DOI: https://doi.org/10.1016/0169-7439(89)80052-8
45. Granitto PM, Furlanello C, Biasioli F, et al. Recursive feature elimination with random forest for PTR-MS analysis of agroindustrial products. Chemometrics and Intelligent Laboratory Systems. 2006; 83(2): 83-90. doi: 10.1016/j.chemolab.2006.01.007 DOI: https://doi.org/10.1016/j.chemolab.2006.01.007
46. Xia J, Psychogios N, Young N, et al. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Research. 2009; 37: W652-W660. doi: 10.1093/nar/gkp356 DOI: https://doi.org/10.1093/nar/gkp356
47. Vapnik V, Cortes C. Support-vector networks. Machine Learning. 1995; 20(3): 273-297. doi: 10.1007/bf00994018 DOI: https://doi.org/10.1007/BF00994018
48. Cortez P, Portelinha M, Rodrigues S, et al. Lamb Meat Quality Assessment by Support Vector Machines. Neural Processing Letters. 2006; 24(1): 41-51. doi: 10.1007/s11063-006-9009-6 DOI: https://doi.org/10.1007/s11063-006-9009-6
49. Pu H, Xie A, Sun DW, et al. Application of Wavelet Analysis to Spectral Data for Categorization of Lamb Muscles. Food and Bioprocess Technology. 2014; 8(1): 1-16. doi: 10.1007/s11947-014-1393-8 DOI: https://doi.org/10.1007/s11947-014-1393-8
50. Donald D, Hancock T, Coomans D, et al. Bagged super wavelets reduction for boosted prostate cancer classification of seldi-tof mass spectral serum profiles. Chemometrics and Intelligent Laboratory Systems. 2006; 82(1-2): 2-7. doi: 10.1016/j.chemolab.2005.08.007 DOI: https://doi.org/10.1016/j.chemolab.2005.08.007
51. Aldoori SKS, Taspinar YS, Koklu M. Distracted Driving Detection with Machine Learning Methods by CNN Based Feature Extraction. International Journal of Applied Mathematics Electronics and Computers. 2021; 9(4): 116-121. doi: 10.18100/ijamec.1035749 DOI: https://doi.org/10.18100/ijamec.1035749
52. Wang J, Li D, Ye Y, et al. A Fluorescent Metal–Organic Framework for Food Real‐Time Visual Monitoring. Advanced Materials. 2021; 33(15). doi: 10.1002/adma.202008020 DOI: https://doi.org/10.1002/adma.202008020
53. Li L, Li M, Liu Y, et al. High-sensitivity hyperspectral coupled self-assembled nanoporphyrin sensor for monitoring black tea fermentation. Sensors and Actuators B: Chemical. 2021; 346: 130541. doi: 10.1016/j.snb.2021.130541 DOI: https://doi.org/10.1016/j.snb.2021.130541
54. Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics. 2020; 21: 1–13. https://doi.org/10.1186/s12864-019-6413-7 DOI: https://doi.org/10.1186/s12864-019-6413-7
55. Deng X, Liu Q, Deng Y, Mahadevan S. An improved method to construct basic probability assignment based on the confusion matrix for classification problem. Information Sciences. 2016 May 1; 340–341: 250–261. https://doi.org/10.1016/j.ins.2016.01.033 DOI: https://doi.org/10.1016/j.ins.2016.01.033




.jpg)
.jpg)

